A group of marine scientists at The Hong Kong University of Science and Technology (HKUST) recently discovered over 7,000 new microbial species in the Oceans, including Acidobacteria – a natural medicinal phylum with the CRISPR gene editing system discovered at sea for the first time, shedding new light on human’s understanding of microbial biodiversity in the oceans and bringing hope to the development of new drugs.
Led by Prof. QIAN Peiyuan, Acting Head of Department of Ocean Science and David von Hansemann Chair Professor of Science at HKUST, the University’s research team collaborated with peers from the King Abdullah University of Science and Technology in Saudi Arabia, University of Georgia in the US and University of Queensland in Australia on sourcing water samples across Pacific, Atlantic and Indian Oceans. Over a span of eight years, the team developed biofilms with the water samples on different materials, eventually discovering more than 7,000 new biofilm-forming species and 10 new bacterial phyla – breaking the existing belief that the world has only 35,000 marine microbial species and 80 bacterial phyla[1]. The finding greatly enhanced human’s knowledge in microbial biodiversity of the oceans.
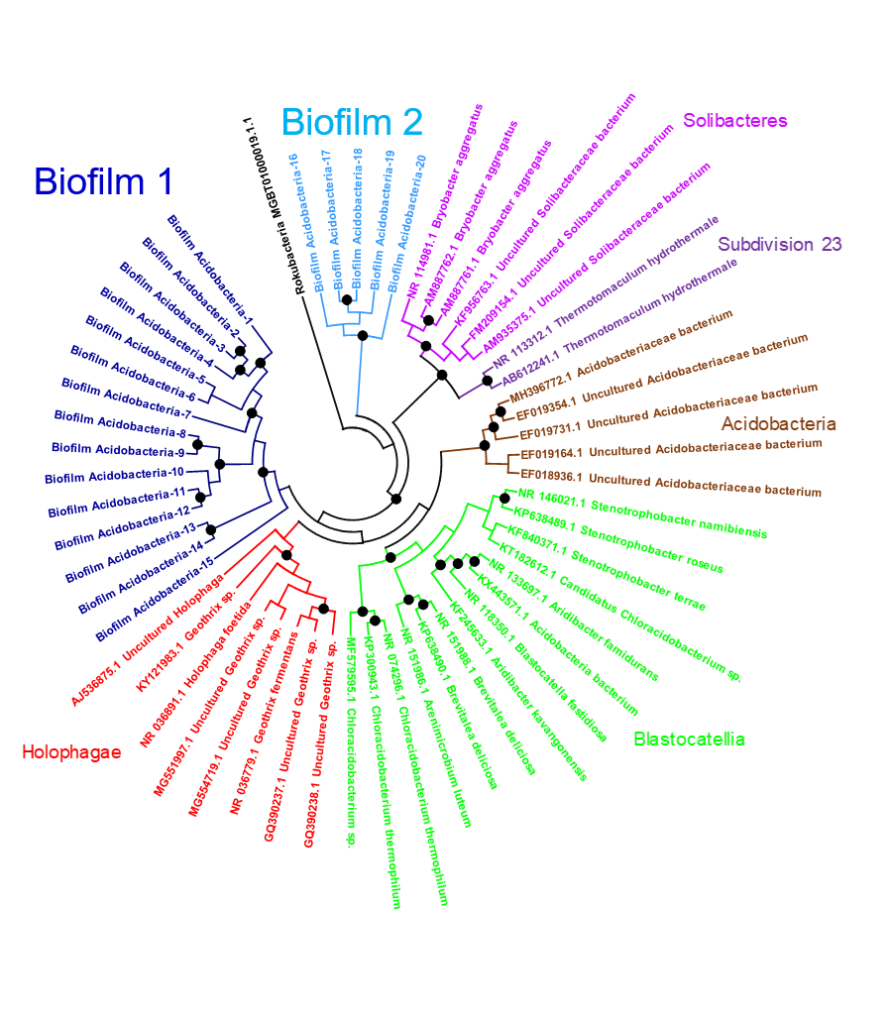
Acidobacteria – one of the new marine phyla identified in this research known only exist in terrestrial soils previously, has been used for developing novel antibiotics and anti-tumor drugs due to its high level of biosynthetic gene clusters. This newly-discovered marine phylum not only shares functions of its terrestrial counterpart, but is also the first ocean species found to contain the CRISPR gene-editing system. The finding was published in the scientific journal Nature Communications.
“The discovery of new marine microbial species has not only improved our understanding of ocean biodiversity, but more importantly, these species have big potential, both in terms of facilitating our understanding of lives and offering new clues to our search of new treatments for diseases,” said Prof. Qian.
CRISPR is a family of DNA sequences found within the genomes of bacterial organisms like Escherichia coli (E-coli); it confers resistance to foreign plasmids or phages and contains gene-editing capabilities. The CRISPR technology is now widely used in agro-industrial and pharmaceutical genetic engineering, for example, to improve yield for soybeans, corn and rice through the creation of more pollution and natural disaster-resistant crops.
Dr. ZHANG Weipeng, a researcher of the team, said the CRISPR system now available in the terrestrial acidobacteria has accuracy issues as it strays from editing target at times. He hoped the new strain could help overcome such shortcomings and bring about more accurate gene editing technology. He added, “Acidobacteria is just one of the many newly-discovered microbial species in this project. With further in-depth study, I am sure there will be more exciting findings to come.”
Prof. Qian is an expert in biofilms. He was awarded the second prize of State Natural Science Award 2016 by The State Council for his important discoveries on how biofilms affect the habitat search of marine benthic animals.
For details, please click here: https://youtu.be/wmDZPXe_RuU
[1] Phylum is one of the units of microorganisms, larger than species with 100 known phyla. The Tara Oceans Project, which is recognized as the most authoritative global ocean microbiome research, found there are 35,000 microbial species in the Oceans.





